![[back]](../left_arrow.gif) |
![[programme]](../up_arrow.gif) |
![[next]](../right_arrow.gif) |

![[back]](../left_arrow.gif) |
![[programme]](../up_arrow.gif) |
![[next]](../right_arrow.gif) |
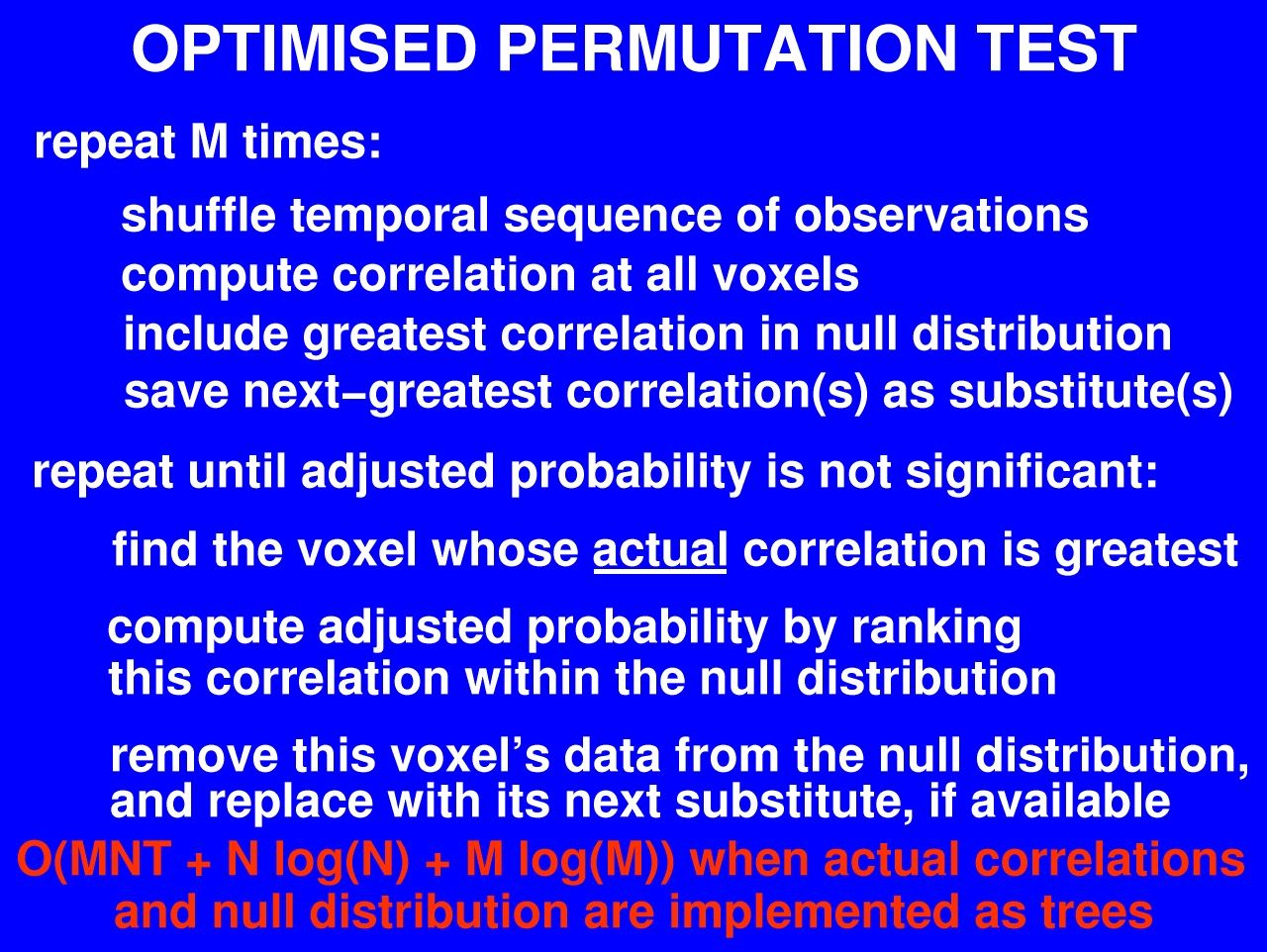
When I first started looking at the problem of significance testing of fMRI data, I thought to myself, there's got to be a better way to do this. So I coded up the original, unmodified step-down algorithm, set it running, and went off to get some coffee. When I came back, the program was still running. So for the rest of the afternoon I worked on other things. And when I checked on it again that evening, it was still running. So I did a few calculations and came up with an estimated time to completion, which worked out to be about a month. And I thought to myself, I'll bet with a month to tinker I can come up with something more efficient.
Actually by the time all the debugging and testing was done it took two months. But it was worth it, I think, since what we have now is an algorithm that runs in nearly linear time -- ten minutes instead of a month. The key difference between this optimised version and the simple step-down algorithm is that when we shuffle the time series and insert the greatest correlation into our null distribution, we also save the next-greatest correlation, and perhaps the next-next-greatest, and so on. We keep these saved values in reserve, and use them to fill in the holes in the distribution when we delete values that have been derived from activated voxels.